Dot chromosome evolution and its role in bird diversification
Understanding how genome architecture shapes biodiversity is a key frontier in evolutionary biology. Among the smallest genomic structures, dot chromosomes (≤ 5 million base pairs) exhibit unique sequences and epigenetic features that distinguish them from larger ones and may encode signatures of evolutionary forces driving speciation. Dot chromosomes have long remained inaccessible to genetic studies and only last year did researchers successfully resolve their structure, thanks to recent advances in long-read sequencing and its application to non-model organisms. For the first time, this breakthrough allows us to investigate the structure and evolutionary significance of dot chromosomes — an opportunity that has eluded researchers until now. This project will build on these advancements to uncover their role in genome evolution and speciation.
Genome architecture behind large white-headed gulls speciation
Understanding how genome structure evolution impacts speciation and lineage diversification is a key focus in current research, powered by high-quality reference genomes, pangenomes, and advanced comparative genomic approaches. Large white-headed gulls (LWHGs) are among the fastest diversifying bird groups, raising questions about how genome structure evolution and trait architecture contribute to rapid reproductive isolation within this clade. In collaboration with the Tree of Life programme at the WSI, Cambridge, UK, I have sequenced and curated high-quality reference genomes for seven gull species. Currently, I am characterising the genomic features of 10 gull species, to address these questions. This representative dataset captures the genetic diversity within LWHGs, enabling an investigation into the relationship between genomic variation (chromosome number and size, gene synteny, transposable element content, and structural variation) and the rapid radiation of this lineage.

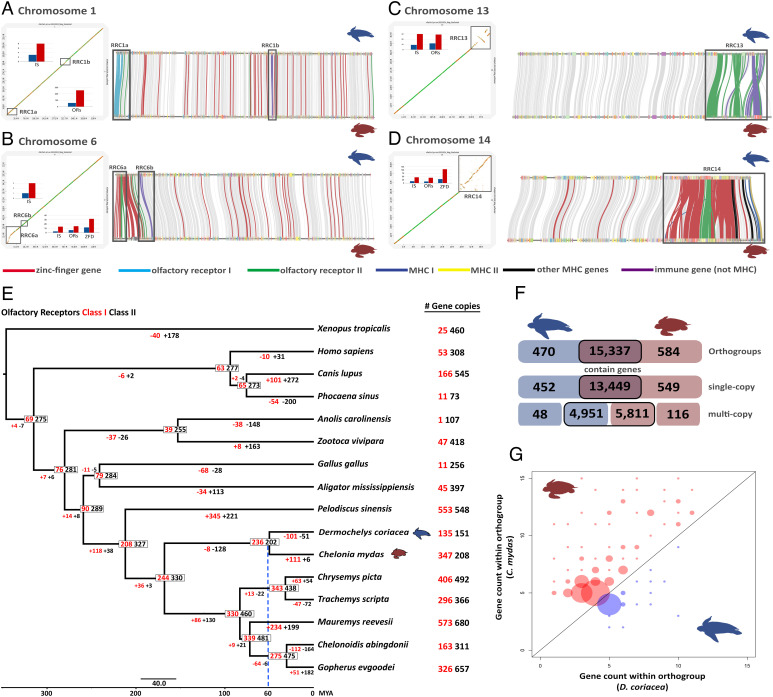
Genomic basis of adaptation in sea turtles
The genomic uniqueness of sea turtles´ molecular evolution was explored using comparative genomics and reverse genetic approaches. We performed a genome-wide analysis of structural variation, chromosome synteny, gene family expansions, and retractions, and a genomic scan for regions affected by natural selection comparing the sea turtles with each other and with non-marine turtles to shed light on sea turtle genomic evolution.
Repeated evolution and molecular basis of adaptation in neotropical freshwater mammals
While the genetic basis of marine adaptations in mammals has been well studied, much less is known about how aquatic mammals have re-adapted to freshwater environments. This transition imposes new osmotic challenges, likely requiring additional changes to the osmoregulatory system. In this study, we investigated 20 genes involved in osmoregulation across strict aquatic mammals—cetaceans and sirenians—that independently colonized freshwater habitats. We found strong signals of positive selection in key genes associated to water homeostasis in the target lineages (Ramos et al. 2023).

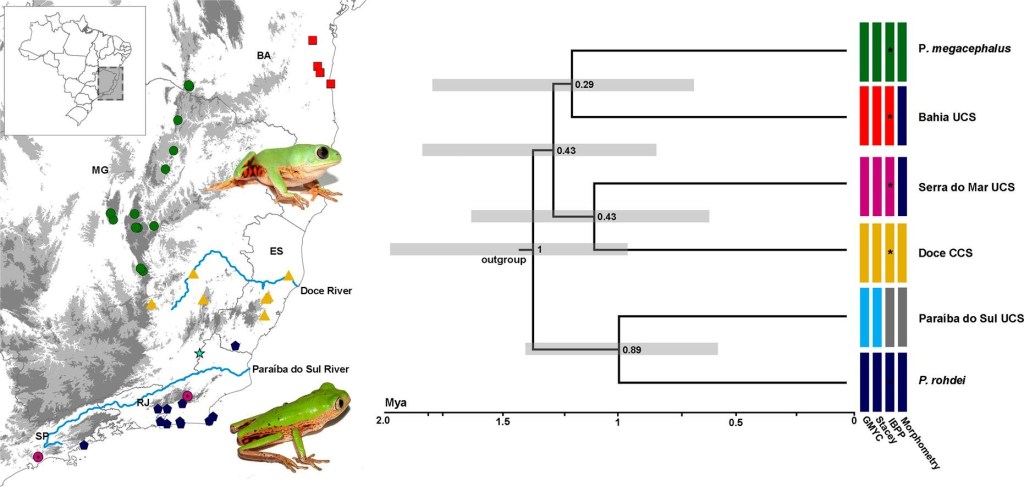
Systematics, phylogeography and conservation of brazilian monkeyfrogs
The Atlantic Forest and the Espinhaço Mountain Range are key biodiversity hotspots in Brazil, home to many endemic species. During my masters, supervised by Dr. Fabrício Rodriguez at LBEM and in collaboration with Dr. Rafael F. Magalhães, at the Federal University of Minas Gerais, I investigate the evolutionary dynamics and conservation needs of two endemic monkey frogs: Pithecopus rohdei from the Atlantic Forest and Pithecopus megacephalus from the Espinhaço Range. By analyzing genetic data from over 140 individuals across 36 locations, we explored population structure, phylogeography, and species delimitation. Our results revealed three distinct lineages within the P. rohdei complex, highlighting the importance of integrative taxonomy in species classification. For P. megacephalus, we identified strong population structuring and evidence of recent expansion in the southern Espinhaço. Given its fragmented distribution in “sky island” habitats, our findings supported its inclusion in the IUCN threatened species list. Overall, this work offers crucial insights into how these populations have evolved and persisted, helping to inform targeted conservation strategies in two of Brazil’s most ecologically important regions.